RESEARCH ARTICLE
- Yonathan Getaneh Yemene 1
- Muluneh Kidane Tufa 2
- Ashenafi Jemal1 Zelalem Getahun2 2
- Bizatu Mengistie 3
1Department Of Emergency Medicine, Saint Paul’s Millennium Medical College, Addis Ababa, Ethiopia
2Department Of Intensive Care Medicine, Saint Paul’s Millennium Medical College, Addis Ababa, Ethiopia
3School of Public Health, Saint Paul’s Millennium Medical College, Addis Ababa, Ethiopia
*Corresponding Author: Muluneh Kidane Tufa
Citation: Muluneh Kidane Tufa2*(2024), Pattern of antimicrobial resistance among patients with culture proven hospital acquired infections at AABET Hospital Addis Ababa, Ethiopia1(1). Journal of Clinical Viral Studies and Microbial Research (JCVSMR) DOI: 10.1875/jcvmr.2024/001
Copyright: © (2024) Muluneh Kidane Tufa2*, this is an open-access article distributed under the terms of The Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: January 22, 2024 | Accepted: May 31, 2024 | Published: June 06, 2024
Abstract
Background: Hospital acquired infection is the most frequently occurring adverse event in any healthcare system regardless of available resources. Studies conducted in Ethiopia reported level of resistance to vancomycin ranging from 8% (Enterococcus species) to 20% (S. aureus)(18). Methods: An institutional based cross-sectional study over a period of one year was conducted by reviewing culture registry logs at AaBET Microbiology lab and cross checking with patient medical records of admissions made to AaBET Hospital from August 1, 2021 to August 1, 2022 . Non probability whole sampling was used on all patients with culture proven HAI who meet the inclusion criteria and who were admitted to AABET Hospital during the study period were. Subsequently data was coded and entered to ep info 7.1 and analysis was conducted using SPSS version 25.0 correlation and bivariant regression was done to assess association. A P- Value of less than 0.05 was taken as significant.
Results- This study shows that the 4 commonest HAI that is diagnosed is surgical site infection (38.3 %) followed by CAUTI (31.2%),HAP(13.5%) and Tracheostomy site infection (6.7%). The most commonly isolated microorganisms were E. Coli(24.07%), Staph Spp. (CONS)(16.4%), Staph Aureus(14.7%), Klebsiella Spp.(14.3%), Pseudomonas Spp.(11.8%),Actinobacter(5.9%),Candida Spp(2.9%). High resistance is observed to Penicillins, Aztreonam & Cephalosporins especially by E. coli & Actinobacter. Additionally prior antimicrobial exposure ,presence of comorbidity, prior healthcare exposure in 90 days, surgery or intubation since admission length of stay prior to diagnosis of hospital acquired infection or total hospital length of stay were not found to be associated with increased resistance in this study.
Conclusion & Recommendation- High resistance is observed to Penicillins, Aztreonam & Cephalosporins especially by E. coli & Actinobacter. With High susceptibility being observed to Amikacin and Chloramphenicol, use of these antimicrobials should be encouraged to susceptible suspected or proven etiologies. Additionally hospital based prospective studies to identify burden of hospital acquired infection and associated risk factors predisposing to antimicrobial resistance and transmission.
Keywords: culture sensitivity and specificity, ventilator associated pneumonia, hospital acquired infection, Surgical Site Infection.
Introduction
An area of public health concern throughout the world is healthcare-associated infection (HAIs), especially those caused by antimicrobial-resistant pathogens. The World Health Organization estimates that HAI is the most frequently occurring adverse event in any healthcare system regardless of available resources (1). It is an infection that occurs after admission to the hospital (48 to 72 hours after admission to 10 -30 days after discharge in some cases); there should not be an active infection at the time of admission and should not be in the incubation period. (2)
Worsening the problem is antibiotic-resistant strains that delay or impede effective treatment. However, the rapid evolution of resistance and the slow discovery of new antimicrobial compounds increasingly reduce treatment options(3).
According to the World Health Organization review, the hospital-wide prevalence of health care-associated infections varies from 5.7% to 19.1%, with a pooled prevalence of 10.1% in low-income countries and prevalence as high as 14.9% reported in centers like Gondar University and Felege Hiwot Hospital (4). Estimates of additional health care costs varied from less than $5(£3;€4) to more than $55000 per patient episode in Europe(5).
A TASH study demonstrate that a laboratory-supported pharmacist-led AMS intervention showed an expected 0.0388 QALYs gained and cost savings of US$82.37 per patient. This translates to 38.8 QALYs gained and total cost savings of US $ 82,370 per 1000 patient admissions(6).
Antimicrobial resistance (AMR) is on the rise worldwide and hospital-wide prevalence of healthcare-associated infections varies from 5.7% to 19.1% (4) Estimates of additional health care cost varied from less than $5(£3;€4) to more than $55000 per patient episode in Europe(7). The risk of hospital-acquired infections increased with invasive devices used. For instance, for treatment and monitoring of patients in Intensive Care Units (ICU) Incidence of device-associated infections (DAIs). [7,8]. The patients identified to have DAI were 5.3% in China,12.2% in Colombia, and 13% in Peruvian ICU patients(8).
It is estimated that the overall short and long-term impact of AMR is likely to be higher in low and middle-income countries (LMICs), especially those in sub-Saharan Africa, mainly owing to a lack of therapeutic options(7) with a pooled prevalence of 10.1% in low-income countries and prevalence as high as 14.9% reported in centers like Gondar University and Felege Hiwot Hospital (4). Associated with this high rates of antimicrobial resistance as demonstrated by a study done in Mekelle, where the resistance rate of E. coli, S. aureus, and S. saprophyticus to ampicillin were 89.0%, 89.0% and 92.3%, respectively(9).
Two major drivers of AMR worldwide include inappropriate antibiotic use in human and animal health and poor infection prevention and control practices(7)In most studies, four major AMR risk factor domains were identified: socio-demographic factors (including migrant status, low income, and urban residence), patient clinical information (includes disease status and certain laboratory results), admission to healthcare settings (includes the length of hospitalization and performance of invasive procedures) and drug exposure (includes current or prior antibiotic therapy) (10). That being said, there is a significant paucity of data regarding the institutional pattern of antimicrobial resistance to help guide treatment and is significantly contributing to patient morbidity, mortality, length of hospital stay, and health care costs. This study is aimed at addressing the pattern of antibiotic resistance and susceptibility which is an important step in the treatment of infections & development of institutional guidelines to better use antibiotics and combat nosocomial infection.
Methods
Study Setting:
This study was conducted at Addis Ababa Burn, Emergency and Trauma /AaBET/ hospital. AaBET hospital is part of Saint Paul's Hospital Millennium Medical College which was established in 2015 and is found in Addis Ababa, Ethiopia. The hospital contains a well-structured Emergency department with 55 beds, which are organized as red, orange, yellow & green areas, an Intensive care unit (ICU) with equipped 11 beds including 03 semi–ICU beds, and 130 beds at inpatient departments of Orthopedics, Neurosurgery, General surgery and burn unit. The hospital provides comprehensive Emergency care 24 hours per day and 7 days per week in Emergency medicine and Critical care, Orthopedic, Neurosurgery, General Surgery and Plastic Surgery.
Study Period:
This study was conducted in patients admitted to Addis Ababa Burn, Emergency and Trauma /AaBET/ hospital from August 1 2020 to August 01/2021, and was conducted from April 15 to October 8,
Study Design:
An institutional-based cross-sectional study over a period of one year will be conducted by reviewing culture registry logs at AABET Microbiology Department and cross-checking with patient medical records of admissions made to AaBET Hospital from August 1, 2021, through August 1, 2022.
Source Population:
• The source population will be all patients admitted in Addis Ababa Burn Emergency and Trauma /AaBET/ hospital during the study Period.
Study Population:
• Patients with Positive culture results and diagnosed with Hospital Acquired Infection (HAP, VAP, CAUTI, Surgical Site Infection, etc…)
INCLUSION AND EXCLUSION CRITERIA
• Inclusion criteria
o All Patients with clinician diagnosis of HAI
o All patients with culture proven HAI
• Exclusion criteria
o Patients without culture result
o Patients whose cultures were sent before 48 hours of health facility stay.
o Patients with incomplete charts or missing charts
Sample Size:
Sampling Procedures: Non Probability Whole sampling was done from all culture logs of cultures sent from AaBET ICU, ED, Wards to the Microbiology lab over a period of one year (from August 1, 2021, through August 1, 2022) that fulfill the inclusion criteria were studied.
Other pertinent data was collected from the patients’ charts using structured data collection instruments.
Data Collection and Procedures:
A structured and pretested data collection tool was formulated for each chart of patients who are diagnosed to have HAI and culture growth and susceptibility pattern will be reviewed and crosschecked with patient chart and the data collection tool was filled by trained data collectors retrospectively from the charts of the study period. Then the questionnaires were checked for completeness and inputted into Ep info 7.1
Data Quality Control and Management:
Collected data was collected, and handled, sorted and inputted by the primary investigator. Data from each data collection tool was reviewed for completeness and indexed charts were reviewed for any missing data.
Data Analysis Plan and Procedures:
The collected data was entered to EPINFO version 3.5.3 and analysis was conducted with SPSS version 25.0. Descriptive statistics like frequencies and cross tabulation were made for most selected variables. Binary logistic regression analysis was used to assess the association between the dependent variable and each independent variable.
Results
Results
Sociodemographic Status
From the 268 cultures sent during the study period, 237 fulfilled the inclusion criteria and were studied. Out of the 237 Patients, 182 (76.8%) were Male and 55 (23.2%) were Female. The mean age of the patients was 31.5 with the youngest studied patient being 3 and the oldest being 87 with the majority (25.3%) of patients in the 31-40 age group. Nearly half (129, 54.4 %) of the patients were from the Oromia region followed by 92 patients (38.8%) from Addis Ababa and 12 patients (5.1%) were from Amhara Region whilst 4 of the remaining patients came from other regions of the country.
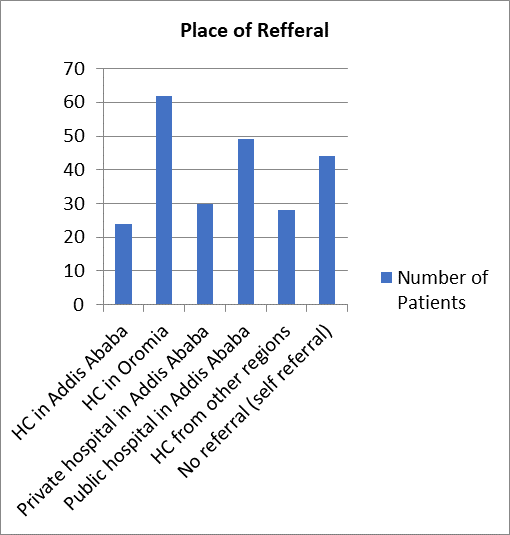
Assessing the referring institution, the majority of the patients (26.2 %) were referred from health centers in Oromia Region as depicted in Figure- 3.
Fig1: Age group of study subjectS
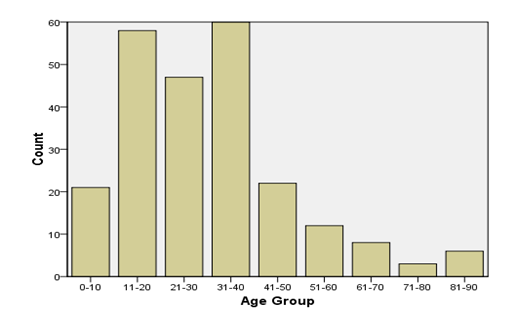
Figure 2- Address of Study Subjects
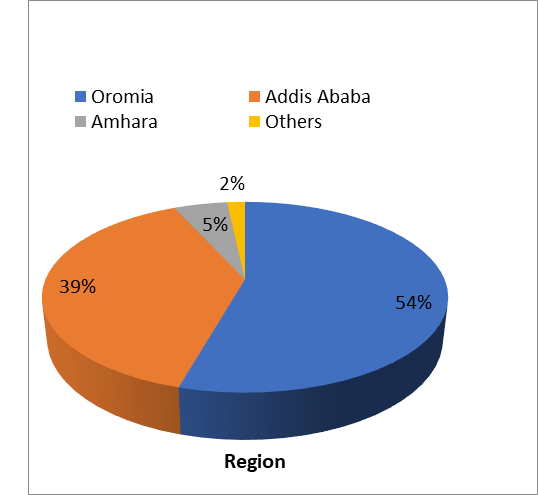
Figure 3- Referring Institution
Classification of patients based on comorbidities and primary disease
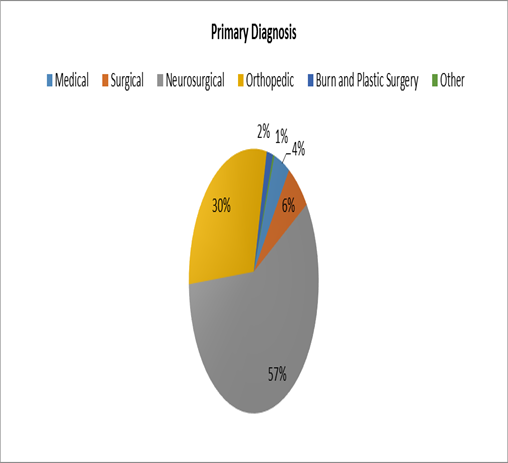
When assessing their primary diagnosis, 136 of the patients (57%) were Neurosurgical, 71 (30%) were orthopedic, 15 (6.3%) were Surgical, 10 were Medical (4.2%), 4 (1.7%) were burn patients and one (0.4 %) was maxillofacial.
Figure 4- Primary Diagnosis
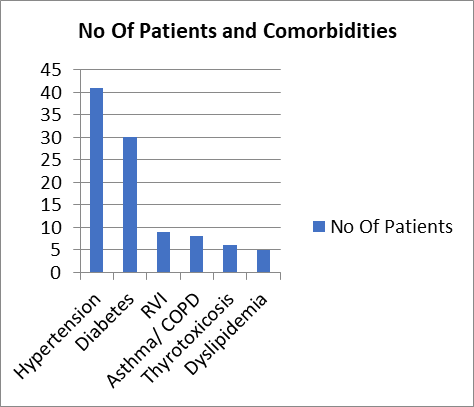
Of the 237 patients studied, 18 had Hypertension, 9 had diabetes, and another 22 patients had both Diabetes and Hypertension. Additionally 5 patients were dyslipidemic and 8 patients had obstructive lung diseases such as Asthma or COPD. The least commonly identified comorbidities were RVI (9) & thyrotoxicosis (6).
Figure 5- Comorbidities
Classification of Patients based on Presumptive Risk factors
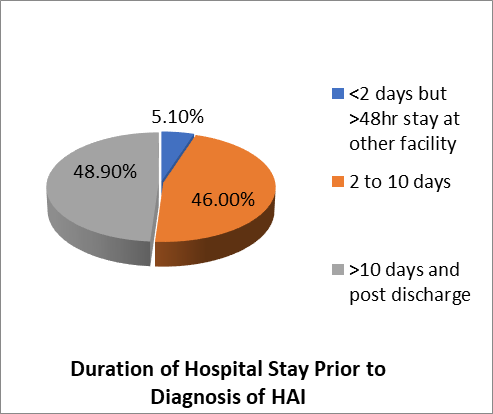
Duration of stay prior to diagnosis of Hospital Acquired Infection
Examination of duration of stay prior to diagnosis of Hospital- acquired infection showed 12 (5.1%) patients were transferred to Aabet Hospital after a stay of at least 48 hours at another facility and were subsequently diagnosed with HAI. 109(46%) Of the Patients stayed 2 to 10 Days prior to diagnosis of HAI whilst 116(48.9%) were diagnosed after 10 days of stay.
Figure 6-Duration of stay prior to diagnosis of HAI
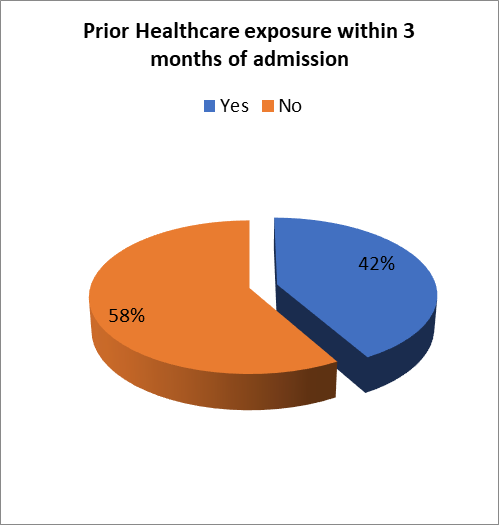
Prior Healthcare exposure within 3 months of admission
Of all the studied patients only 99 (41.8%) had prior healthcare exposure within 3 months of presentation.
Figure 7- Prior Healthcare Exposure within 3 months
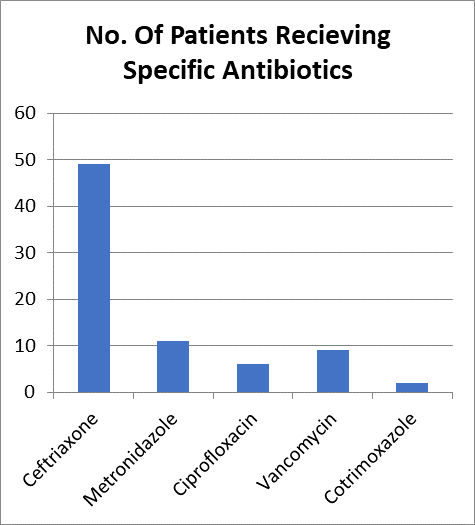
Percentage of patients with prior antimicrobial exposure
From those studied only 75 (31.6 %) of the patients had confirmed prior antimicrobial Exposure where 47(62.6%) of them received Ceftriaxone, 11(14%) Received Metronidazole and an additional 2 patients received both Ceftriaxone and Metronidazole. Additionally 6 (8%) had received Ciprofloxacin, 9 (12%) received Vancomycin & 2 (2%) received cotrimoxazole.
Figure 8-No. Of Patients Receiving Specific Antibiotics Prior to Presentation
Patient who underwent invasive Procedures
From those Patients studied 173 (73 %) of them had underwent Surgery and one of the patients had a central vascular catheter inserted.
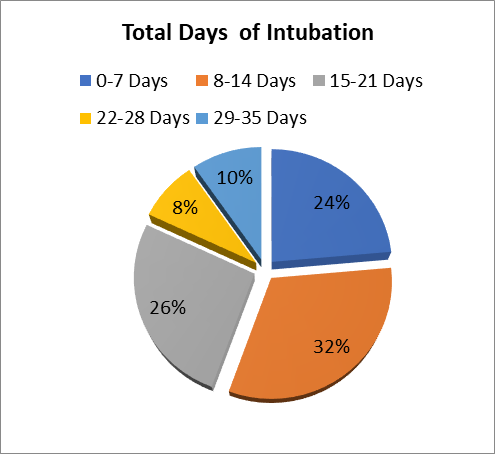
A total of 79 patients had been admitted to the ICU and 7 of these patients were readmitted after disposition. Endotracheal Intubation was performed in 72 Patients and the majority (31.9%) were intubated for 8 to 14 days with maximum intubation duration of 35 Days. From all Patients Studied the minimum length of Hospital stay was 3 days and the longest was 109 days with a mean length of stay of 29.8 days.
Figure 9- Total Hospital Length of Stay
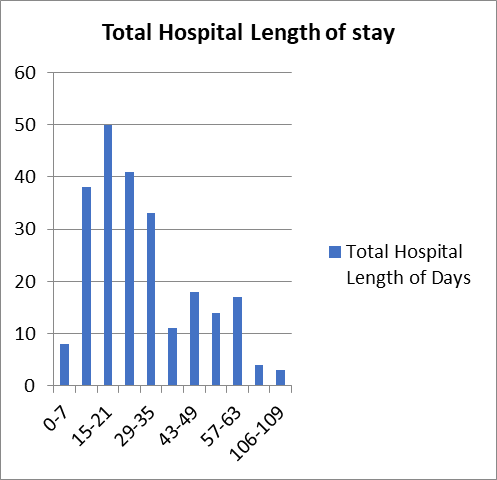
Figure 10- Total Days of Intubation
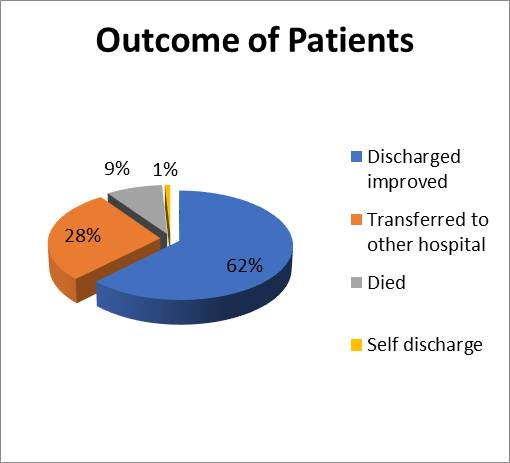
Final Outcome of Patients
Of all the patients studied 148 (62.4%) were discharged with improvement and 21 (8.8%) died and 2(0.8%) went against medical advice, the remaining 66 (27.8%) patients were transferred to other hospitals.
Figure 11- Final Outcome of Patients
Type of HAI
The most common HAI identified in this study was wound site infection, followed by CAUTI, HAP, tracheostomy site infection, and VAP. Other less common infections are post-traumatic meningitis/ventriculitis and central line-related bloodstream infections.
Table 1- Types of HAI
| Frequency | Percent | |
| HAP | 32 | 13.5% |
| VAP | 14 | 5.9% |
| CAUTI | 74 | 31.2% |
| Surgical site infection | 91 | 38.4% |
| Central line associated bloodstream infections | 1 | 0.4% |
| Tracheostomy Site Infection | 16 | 6.8% |
| Post Traumatic Meningitis/ Ventriculitis | 8 | 3.4% |
The most common growth in urine cultures was E.Coli(36) Followed by Klebsiella Spp. (17), actionbacter(8), Pseudomonas (7), Candida Albicans (7), Staph spp. (5), Enterobacter spp. , S. Aureus (3), Citrobacter (2),Proteus (2), and Enterococcus respectively. The commonest Organisms identified in blood cultures were Staph Spp.(CONS) , Staph Aureus (9),Klebsiella Spp. (6), E. Coli (3), Pseudomonas (3), Actinobacter (2), Citrobacter, Enterobacter, Proteus, respectively
Table 2 - Comparison of source of culture and Microorganism identified
| Organism | Positive Culture Source |
Total | ||||||||||||
| Urine culture | Blood Culture | CSF culture | Tracheal aspirate | Wound culture | Pleural fluid | |||||||||
| Klebsiella spp | 17 | 6 | 0 | 1 | 9 | 1 | 34 | |||||||
| Psuedomonas spp | 7 | 3 | 2 | 8 | 8 | 0 | 28 | |||||||
| Staph Aureus | 3 | 9 | 1 | 3 | 19 | 0 | 35 | |||||||
| Staph spp (CONS) | 5 | 17 | 1 | 1 | 14 | 1 | 39 | |||||||
| Acinetobacter | 8 | 2 | 1 | 0 | 3 | 0 | 14 | |||||||
| Citerobacter | 2 | 1 | 0 | 1 | 2 | 0 | 6 | |||||||
| E coli | 36 | 4 | 2 | 2 | 12 | 1 | 57 | |||||||
| Enterobacter spp | 4 | 1 | 1 | 1 | 3 | 0 | 10 | |||||||
| Candida albicans | 7 | 0 | 0 | 0 | 0 | 0 | 7 | |||||||
| Proteus | 2 | 1 | 0 | 0 | 3 | 0 | 6 | |||||||
| Enterococcus | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |||||||
| Total | 92 | 44 | 8 | 17 | 73 | 3 | 237 | |||||||
The etiologies responsible for CAUTI which is the second most common HAI in this study are E. Coli, Klebsiella spp, Staph. Spp, Pseudomonas, Staph Aureus in descending order of frequency.
When assessing the culprits for HAP Klebsiella spp., E.Coli, Staph spp. (CONS), Pseudomonas , Actinobacter, Staph Aureus, Enterobacter spp, Citrobacter, Proteus, in descending order of frequency, were identified. Those with Suspected VAP had culture growth showing E.Coli, Pseudomonas, Staph Spp., klebsiella spp, Actinobacter , Enterobacter spp.
Table 3- Comparison of Identified Microorganism and Clinical Diagnosis

Pattern of Antimicrobial Resistance
Of the 237 patients studied, 145 were tested for cephalosporin resistance, and 102 (70.3%) samples showed resistance, 42 (29.6%) samples were susceptible, and 1 sample was indeterminate. The most resistant organisms in descending order of frequency were E. Coli (37,36.2%) , Klebsiella (23,22.5%), Staph spp(CONS) (10,0.98%),Actinobacter (9,8.8%). Staph Aureus (7,6.8%) .
Additionally from all cultures collected 123 were tested for resistance against Aminoglycosides (i.e. either Tobramycin or Gentamycin) and it showed 69 of the organisms were sensitive whilst 43 were resistant and the remaining 11 were indeterminate. The Organism showing the most resistance to aminoglycosides was E. Coli (13, 30.2%) followed by Staph spp (CONS)(10,23.2%) , Actinobacter (6,13.9%) and Klebsiella (5,11.6%). On the contrary sensitivity to this group of antibiotics was observed to be highest among S. Aureus(27,39.1%) followed by Staph spp. (CONS)(16,23.1%), Pseudomonas(9,13.04%) and E.Coli(8,11.5%). However less amount of resistance is observed among the 90 samples tested for resistance against Amikacin, 81 (90%) of the samples were sensitive while 4(4.4%) showed resistance and 5(5.5%) were indeterminate.
When assessing the degree of penicillin group resistance among the 86 samples tested, 70 (81.3%) of the samples showed resistance, 13 (15.1%) were sensitive whilst 3(3.4%) were indeterminate. The Highest resistance to Penicillins was seen in E. Coli where 25 (89.2%) of the organisms were resistant while Klebsiella spp. 12(92.3%) , Pseudomonas Species 9(52%), S. Aureus 7 (70%) , Staph spp 6(85.7%) ,Citrobacter 5(100%).
Regarding resistance to Fluoroquinolones (Ciprofloxacin and Norfloxacin) 108 samples were tested and 50 (46.2%) of the samples showed resistance while 51 (47.2%) were sensitive and 7 were indeterminate. The highest resistance was seen among E.Coli(21, 61.7%) followed by Klebsiella (16,72.7%), Staph spp(CONS)(3,27%). The Highest sensitivity was seen by S. Aureus(12,85.7%),Pseudomonas (9,81.8%), Staph spp. (CONS)(8,72.2%)
When assessing resistance to Meropenem from the 98 tested samples 78(79.5%) were sensitive while 18(18.3%) were resistant and 2 (2%) were indeterminant. The highest resistance was seen in Klebsiella (6, 24%) followed by Actinobacter(5,45.4%) and Pseudomonas spp. (4,17.3%). The sensitivity of organisms in desending order was E.Coli (26,92.8%), followed by Pseudomonas (19,82.6%),Klebsiella (18,72%),Enterobacter (7,87.5%),Actinobacter (5,45.5%) and proteus (2,100%).
Chloramphenicol was tested on 39 samples and 25 of the samples showed sensitivity while 14 were resistant, the highest resistance was seen by Klebsiella(4,50%) followed by E.Coli(3,18.7%), Proteus (2,100%), Citrobacter (2,100%). While Sensitivity was seen by E. Coli (13,81.2%),Staph Aureus (5,100%), Klebsiella (4,50%). The greatest resistance was observed on aztreonam, where all 25 samples showed resistance, with the greatest resistance seen by Staph Aureus, Staph spp. (CONS) and E.coli.
The other group of antimicrobials studied was Trimethoprim-Sulfamethoxazole, 96 samples were tested and 63(65.6%) showed resistance while 30(31.2%) showed sensitivity and 3(3.1%) were indeterminate. The Most common microbe showing resistance was E. Coli (24, 7
7.4%) followed by Staph spp. (CONS)(16,80%) followed by Klebsiella spp(8,80%), S. Aureus (6,31.5%), Citrobacter (4,100%), Actinobacter (3,42%). The highest sensitivity was seen by S.Aureus (12, 63.1%), followed by E.Coli(7,22.5%), Staph spp. (CONS)(4,20%).
Table 4 - Comparison of Identified Organism and Pattern of Susseptability to Major Groups of Antimicrobials
| Specific_microbe | Total | ||||||||||
| Klebsiella spp | Psuedomonas spp | Staph. Aureus | Staph spp. (CONS) | Acinetobacter | Citerobacter | E coli | Enterobacter spp | Proteus | |||
| Cephalosporins | Indeterminate | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Sensitive | 1 | 6 | 13 | 13 | 0 | 1 | 6 | 1 | 1 | 42 | |
| Resistant | 23 | 2 | 7 | 10 | 9 | 4 | 37 | 5 | 5 | 102 | |
| Total | 24 | 8 | 20 | 23 | 9 | 5 | 44 | 6 | 6 | 145 | |
| Aminoglycosides | Indeterminate | 1 | 0 | 1 | 1 | 1 | 0 | 6 | 1 | 0 | 11 |
| Sensitive | 5 | 9 | 27 | 16 | 2 | 0 | 8 | 1 | 1 | 69 | |
| Resistant | 5 | 2 | 2 | 10 | 6 | 2 | 13 | 1 | 2 | 43 | |
| Total | 11 | 11 | 30 | 27 | 9 | 2 | 27 | 3 | 3 | 123 | |
| Fluoroquinolones | Indeterminate | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 0 |
| Sensitive | 6 | 9 | 12 | 8 | 4 | 3 | 7 | 2 | 0 | 0 | |
| Resistant | 16 | 2 | 2 | 3 | 2 | 0 | 21 | 1 | 2 | 1 | |
| Total | 22 | 11 | 14 | 11 | 6 | 3 | 34 | 4 | 2 | 1 | |
| Meropenem | Indeterminate | 1 | 0 | - | - | 1 | 0 | 0 | 0 | 0 | 2 |
| Sensitive | 18 | 19 | - | - | 5 | 1 | 26 | 7 | 2 | 78 | |
| Resistant | 6 | 4 | - | - | 5 | 0 | 2 | 1 | 0 | 18 | |
| Total | 25 | 23 | - | - | 11 | 1 | 28 | 8 | 2 | 98 | |
| Trimethoprim Sulfamethoxazole | Indeterminate | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 3 |
| Sensitive | 2 | 0 | 12 | 4 | 2 | 0 | 7 | 2 | 1 | 30 | |
| Resistant | 8 | 1 | 6 | 16 | 3 | 4 | 24 | 0 | 1 | 63 | |
| Total | 10 | 1 | 19 | 20 | 7 | 4 | 3 | 2 | 2 | 96 | |
| Penicilins | Indeterminate | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 |
| Sensitive | 1 | 7 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 13 | |
| Resistant | 12 | 9 | 7 | 6 | 1 | 5 | 25 | 2 | 3 | 70 | |
| Total | 13 | 17 | 10 | 7 | 1 | 5 | 28 | 2 | 3 | 86 | |
| Amikacin | Indeterminate | 0 | 0 | - | - | 0 | 0 | 5 | 0 | 0 | 5 |
| Sensitive | 28 | 20 | - | - | 6 | 2 | 17 | 7 | 1 | 81 | |
| Resistant | 1 | 1 | - | - | 0 | 0 | 0 | 1 | 1 | 4 | |
| Total | 29 | 21 | - | - | 6 | 2 | 22 | 8 | 2 | 90 | |
| Aztreonam | Indeterminate | - | - | 15 | 8 | - | - | 2 | - | - | 25 |
| Sensitive | - | - | - | - | - | - | - | - | - | - | |
| Resistant | - | - | - | - | - | - | - | - | - | - | |
| Total | - | - | 15 | 8 | - | - | 2 | - | - | 25 | |
| Chloramphenicol | Indeterminate | - | - | - | - | - | - | - | - | - | - |
| Sensitive | 4 | 0 | 5 | 1 | 0 | 0 | 13 | 2 | 0 | 35 | |
| Resistant | 4 | 1 | 0 | 1 | 1 | 2 | 3 | 0 | 2 | 14 | |
| Total | 8 | 1 | 5 | 2 | 1 | 2 | 16 | 2 | 2 | 39 | |
Table 5 - Comparison of Identified Organism and Pattern of Susceptibility to Antimicrobials
| Specific_microbe | Total | ||||||||||
| Klebsiella spp | Psuedomonas spp | Staph. Aureus | Staph spp. (CONS) | Acinetobacter | Citerobacter | E coli | Enterobacter spp | Proteus | |||
| Amikacin | Indeterminate | 0 | 0 | - | - | 0 | 0 | 5 | 0 | 0 | 5 |
| Sensitive | 28 | 20 | - | - | 6 | 2 | 17 | 7 | 1 | 81 | |
| Resistant | 1 | 1 | - | - | 0 | 0 | 0 | 1 | 1 | 4 | |
| Total | 29 | 21 | - | - | 6 | 2 | 22 | 8 | 2 | 90 | |
| Aztreonam | Indeterminate | - | - | 15 | 8 | - | - | 2 | - | - | 25 |
| Sensitive | - | - | - | - | - | - | - | - | - | - | |
| Resistant | - | - | - | - | - | - | - | - | - | - | |
| Total | - | - | 15 | 8 | - | - | 2 | - | - | 25 | |
| Cefazolin | Indeterminate | 0 | - | - | - | - | - | 1 | 0 | 0 | 1 |
| Sensitive | - | - | - | - | - | - | - | - | - | - | |
| Resistant | 2 | - | - | - | - | -- | 2 | 1 | 1 | 6 | |
| Total | 2 | - | - | - | - | - | 3 | 1 | 1 | 7 | |
| Cefepime | Indeterminate | - | - | - | - | - | - | -- | - | - | - |
| Sensitive | 0 | 4 | - | - | 0 | 0 | 1 | - | 0 | 5 | |
| Resistant | 5 | 0 | - | - | 2 | 1 | 9 | - | 2 | 19 | |
| Total | 5 | 4 | - | - | 2 | 1 | 10 | - | 2 | 24 | |
| Cefoxitin | Indeterminate | - | - | - | - | - | - | - | - | - | - |
| Sensitive | - | - | 14 | 13 | - | - | 1 | - | - | 28 | |
| Resistant | - | - | 7 | 10 | - | - | 0 | - | - | 17 | |
| Total | - | - | 21 | 23 | - | - | 1 | - | - | 45 | |
| Cefotaxime | Indeterminate | - | - | - | - | - | - | - | - | - | - |
| Sensitive | 0 | - | - | - | 0 | - | 3 | - | - | 3 | |
| Resistant | 3 | - | - | - | 2 | - | 4 | - | - | 9 | |
| Total | 3 | - | - | 2 | - | 7 | - | - | 12 | ||
| Ceftazidime | Indeterminate | 1 | 0 | - | - | 0 | - | 1 | - | - | 2 |
| Sensitive | 1 | 4 | - | - | 0 | - | 0 | - | - | 6 | |
| Resistant | 6 | 2 | - | - | 4 | - | 7 | - | - | 19 | |
| Total | 8 | 6 | - | - | 4 | - | 8 | - | - | 27 | |
| Ceftazidime Avibactam | Indeterminate | - | - | - | - | - | - | - | - | - | |
| Sensitive | - | - | - | - | - | - | 1 | - | 1 | 2 | |
| Resistant | - | - | - | - | - | - | - | - | - | ||
| Total | - | - | - | - | - | - | 1 | - | 1 | 2 | |
| Ceftriaxone | Indeterminate | 0 | - | - | - | 0 | 0 | 1 | 0 | 0 | 1 |
| Sensitive | 1 | - | - | - | 0 | 1 | 2 | 0 | 1 | 5 | |
| Resistant | 13 | - | - | - | 5 | 3 | 24 | 5 | 3 | 53 | |
| Total | 14 | - | - | - | 5 | 4 | 27 | 5 | 4 | 59 | |
| Cefuroxime | Indeterminate | 0 | - | - | - | - | 0 | 1 | 0 | - | 1 |
| Sensitive | 0 | - | - | - | - | 0 | 1 | 0 | - | 1 | |
| Resistant | 1 | - | - | - | - | 1 | 7 | 1 | - | 10 | |
| Total | 1 | - | - | - | - | 1 | 9 | 1 | - | 12 | |
| Chlorampenicol | Indeterminate | - | - | - | - | - | - | - | - | - | |
| Sensitive | 4 | 0 | 5 | 1 | 0 | 0 | 13 | 2 | 0 | 25 | |
| Resistant | 4 | 1 | 0 | 1 | 1 | 2 | 3 | 0 | 2 | 14 | |
| Total | 8 | 1 | 5 | 2 | 1 | 2 | 16 | 2 | 2 | 39 | |
| Ciprofloxacin | Indeterminate | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 0 | 6 |
| Sensitive | 5 | 7 | 10 | 6 | 3 | 2 | 7 | 2 | 0 | 42 | |
| Resistant | 8 | 2 | 2 | 3 | 2 | 0 | 15 | 1 | 2 | 35 | |
| Total | 13 | 9 | 12 | 9 | 5 | 2 | 27 | 4 | 2 | 83 | |
| Norfloxacin | Indeterminate | - | - | - | - | - | - | 1 | - | - | 1 |
| Sensitive | 1 | 2 | 4 | 3 | 1 | 1 | 1 | 0 | - | 13 | |
| Resistant | 8 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | - | 15 | |
| Total | 9 | 2 | 4 | 3 | 1 | 1 | 8 | 1 | - | 29 | |
| Clindamycin | Indeterminate | - | - | 0 | 2 | - | - | 0 | - | - | 2 |
| Sensitive | - | - | 24 | 12 | - | - | 1 | - | - | 37 | |
| Resistant | - | - | 0 | 4 | - | - | 0 | - | - | 4 | |
| Total | - | - | 24 | 18 | - | - | 1 | - | - | 43 | |
| Gentamycin | Indeterminate | 1 | 0 | 1 | 1 | 1 | 0 | 4 | 1 | 0 | 9 |
| Sensitive | 5 | 7 | 11 | 7 | 2 | 0 | 7 | 1 | 1 | 41 | |
| Resistant | 3 | 2 | 2 | 8 | 6 | 2 | 11 | 1 | 2 | 37 | |
| Total | 9 | 9 | 14 | 16 | 9 | 2 | 22 | 3 | 3 | 87 | |
| Tobramycin | Indeterminate | 0 | 0 | - | - | - | - | 2 | - | - | 2 |
| Sensitive | 0 | 2 | - | - | - | - | 1 | - | - | 3 | |
| Resistant | 2 | 0 | - | - | - | - | 2 | - | - | 4 | |
| Total | 2 | 2 | - | - | - | - | 5 | - | - | 9 | |
| Meropenem | Indeterminate | 1 | 0 | - | - | 1 | 0 | 0 | 0 | 0 | 2 |
| Sensitive | 18 | 19 | - | - | 5 | 1 | 26 | 7 | 2 | 78 | |
| Resistant | 6 | 4 | - | - | 5 | 0 | 2 | 1 | 0 | 18 | |
| Total | 25 | 23 | - | - | 11 | 1 | 28 | 8 | 2 | 98 | |
| Nalidixic Acid | Indeterminate | - | - | - | - | - | - | - | - | - | - |
| Sensitive | 1 | - | - | - | 1 | 0 | 5 | 0 | 1 | 8 | |
| Resistant | 0 | - | - | - | 1 | 1 | 17 | 2 | 1 | 22 | |
| Total | 1 | - | - | - | 2 | 1 | 22 | 2 | 2 | 30 | |
| Nitrofuranroin | Indeterminate | 0 | - | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Sensitive | 5 | - | 3 | 2 | 0 | 0 | 24 | 1 | 0 | 35 | |
| Resistant | 2 | - | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 9 | |
| Total | 7 | - | 3 | 3 | 2 | 1 | 25 | 2 | 2 | 45 | |
| Ampicillin | Indeterminate | 0 | 0 | - | - | - | 0 | 2 | 0 | 0 | 2 |
| Sensitive | 0 | 0 | - | - | - | 0 | 1 | 0 | 0 | 1 | |
| Resistant | 11 | 1 | - | - | - | 5 | 24 | 2 | 3 | 46 | |
| Total | 11 | 1 | - | - | - | 5 | 27 | 2 | 3 | 49 | |
| Penicillin | Indeterminate | - | - | - | - | - | - | - | - | ||
| Sensitive | 0 | - | 3 | 1 | - | - | 0 | - | - | 4 | |
| Resistant | 1 | - | 7 | 6 | - | - | 1 | - | - | 15 | |
| Total | 1 | - | 10 | 7 | - | - | 1 | - | - | 19 | |
| Piperacillin | Indeterminate | - | 4 | - | - | - | - | - | - | - | 4 |
| Sensitive | - | 2 | - | - | - | - | - | - | - | 2 | |
| Resistant | - | 6 | - | - | - | - | - | - | - | 6 | |
| Total | - | 12 | - | - | - | - | - | - | - | 12 | |
| Piperacillin Tazobactam | Indeterminate | - | - | - | - | - | - | - | - | ||
| Sensitive | 1 | 6 | - | - | 0 | - | - | - | - | 7 | |
| Resistant | 0 | 9 | - | - | 1 | - | - | - | - | 10 | |
| Total | 1 | 15 | - | - | 1 | - | - | - | - | 17 | |
| Tetracycline | Indeterminate | 0 | - | 0 | 0 | 0 | - | 1 | 0 | - | 1 |
| Sensitive | 8 | - | 2 | 0 | 1 | - | 3 | 3 | - | 17 | |
| Resistant | 4 | - | 1 | 2 | 0 | - | 6 | 1 | - | 14 | |
| Total | 12 | - | 3 | 2 | 1 | - | 10 | 4 | - | 32 | |
| Trimethoprim Sulfamethoxazole | Indeterminate | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 3 |
| Sensitive | 2 | 0 | 12 | 4 | 2 | 0 | 7 | 2 | 1 | 30 | |
| Resistant | 8 | 1 | 6 | 16 | 3 | 4 | 24 | 0 | 1 | 63 | |
| Total | 10 | 1 | 19 | 20 | 7 | 4 | 31 | 2 | 2 | 96 | |
A correlation was observed between age group and the presence of resistant microorganisms with Pearson Chi-Square of 0.05 and a bivariate logistic regression was performed, no association was observed (P-value-0.23). Additionally, a weak observed correlation between prior antimicrobial exposure and the presence of resistance was noted (Pearson Chi-Square of 0.06) and was found to have a weak association on bivariant logistic regression with P value=0.069, AOR=1.9, CI-0.949-4.096). There was a correlation observed between the presence of a central vascular catheter and the Presence of a Resistant organism with Pearson Chi-Square of 0.059 but no association was observed (P-value-1.0). However, there was no correlation between Bacterial Resistance and the presence of comorbidity, prior healthcare exposure in 90 days, surgery, or intubation since admission, or total hospital length of stay.
Discussion
This study shows that the 4 commonest HAI that is diagnosed is Wound site infection (38.3 %) followed by CAUTI (31.2%), HAP (13.5%) and Tracheostomy site infection (6.7%). However, the prevalence of SSI in this study is higher than in one study done in Ghana where it constituted 14-31% of all hospital-acquired infections. (13) This may be attributed to the fact that the study was done at a trauma center where majority of patients have sustained contaminated wounds or undergone surgical procedures. The commonest isolated microorganisms in descending order of frequency were E. Coli (24.07%) followed by Staph spp. (CONS)(16.4%), Staph Aureus (14.7%), Klebsiella spp.(14.3%), Pseudomonas spp.(11.8%) & Actinobacter (5.9%) as Compared to a multicentric study done in Amhara region which found the highest isolated microorganisms Klebsiella spp. (22.44%) and Staphylococcus Aureus (20.4%). (22)
When assessing the resistance patterns of the commonly isolated pathogens E. Coli showed a high rate of resistance to different classes of antibiotics including Aztreonam (100%), Penicillins(89.2%), cephalosporin’s (36.2 %), Aminoglycosides (30.2 %) and Fluoroquinolones(61.7%) which shows a similar state of resistance as compared to the previous reported resistance of 52-92% to Penicillins 57-84% to cephalosporin’s and 56-78% to fluoroquinolones. However high rates of susceptibility were noted to Meropenem (92.8%), Chloramphenicol (81.2%), Amikacin (77%).
The Second most common organism isolated was staph spp. (CONS) which showed resistance of 72 % to Cotrimoxazole, 43% to cephalosporin’s ,37% Aminoglycosides showed the highest sensitivity to Clindamycin and Ciprofloxacin at 66?ch. this shows an alarming increase in resistance in this organism to Cotrimoxazole as compared to a previous pooled study which showed an 11% resistance. (20)
The resistance of Staphylococcus Aureus was highest to Aztreonam at 100%, Penicillin group of antimicrobial at 70 %, This finding shows a lesser degree of resistance as compared to a Kenyan study which showed 97% resistance to this group of antimicrobial. In this study it also showed significant resistance to Cotrimoxazole (31.5%) which is much less as compared to the previous study where 70% of the isolates were resistant. However the organisms showed excellent susceptibility to Chloramphenicol (100%) and Fluoroquinolones (85.7%). (18)
Klebsiella spp. which is the most incriminated organism as the cause of HAP in this study showed the most resistance to Penicillins at 92.3 %, Cotrimoxazole (80 %) Fluoroquinolones (72.7%), Meropenem (24%) cephalosporin’s at 22.5%. However higher rates of susceptibility were observed to Amikacin (96 %), Meropenem (72%), Chloramphenicol (50%) and Other Aminoglycosides (45%). Tough this study more or less shows a similar resistance of Klebsiella spp to penicillin’s as the study done in Jimma University hospital where it was 100% resistant to Ampicillin and Amoxicillin , the sensitivity of the organism to Ciprofloxacin is 38% as compared to the aforementioned study where the sensitivity was 100%. (20)
Pseudomonas Resistance was highest on this study to cotrimoxazole and Chloramphenicol (100?ch) likely due to the fact only one sample was tested for each and found to be resistant. Penicillin resistance was at 52% and 18 % to Fluoroquinolones. The highest sensitivity was observed to Amikacin 95%, Meropenem 82%, 81% Fluoroquinolones, 81% OtherAminoglycosides. A Higher susceptibility to Meropenem (82%) was noted in our study as compared to a study done in Kenyatta National Hospital (KNH) in Nairobi, Kenya. %). (18)
Acinetobacter spp. Was Found to have 100% resistance to cephalosporins, Penicillins, and Chloramphenicol, the latter 2 however were tested on one sample and may not be representative. The microbe was also found to have 66% resistance to Aminoglycosides (Gentamycin & Tobramycin) & 42% to Cotrimoxazole, However, it showed Full susceptibility to Amikacin and 66% to Fluoroquinolones and 45 % to Meropenem.
Overall there is a concerning increase in the resistance of E. Coli, Staph spp. (CONS), Actinobacter. An increase in antibiotic resistance amongst these pathogens will make it difficult to treat the infections with locally available antibiotics.
Finally, in assessing possible risk factors for antimicrobial resistance, there was a correlation observed between an increase in age, the presence of a central vascular catheter, and the presence of resistant organisms however no association was found. Additionally, prior antimicrobial exposure, presence of comorbidity, prior healthcare exposure in 90 days, surgery or intubation since admission length of stay before the diagnosis of HAI, or total hospital length of stay were not found to be associated with increased resistance in this study. Acinetobacter spp. was Found to have 100% resistance to cephalosporins, Penicillins, and Chloramphenicol, the latter two however were tested on one sample and may not be representative. The microbe was also found to have 66% resistance to Aminoglycosides (Gentamycin & Tobramycin) & 42% to Cotrimoxazole, However, it showed full susceptibility to Amikacin and 66% to Fluoroquinolones and 45 % to Meropenem.
Conclusion
This study shows that the 4 commonest HAIs that are diagnosed are surgical site infections followed by CAUTI, HAP, and Tracheostomy site infections. The most commonly isolated microorganisms were E. Coli, Staph Spp. (CONS), Staph Aureus, Klebsiella Spp., Pseudomonas Spp.,Actinobacter & Candida Spp. A Staggering resistance is observed to Penicillins, Aztreonam & Cephalosporins especially by E.Coli and Actinobacter. The Commonest Etiologies identified have good susceptibility to Meropenem, Amikacin, Chloramphenicol, and to a certain extent Fluoroquinolones. Therefore, treatment and management of HAIs should be based on knowledge of bacterial etiology and antibiotic resistance patterns. Additionally, prior antimicrobial exposure, presence of comorbidity, prior healthcare exposure in 90 days, surgery or intubation since admission length of stay before the diagnosis of HAI, or total hospital length of stay were not found to be associated with increased microbial resistance in this study.
Acronyms and Abbreviations
AICU: Adult Intensive Care Unit
AKI: Acute Kidney Injury
CKD: Chronic Kidney Disease
CRRT: Continuous Renal Replacement Therapy
IHM: In Hospital Mortality
ISS: Injury Severity Score
KDIGO: Kidney Disease Improving Global Outcomes
LOS: Length of Stay
RRT: Renal Replacement Therapy
SCR: Serum Creatinine
Declarations
Consent for publication
Participants consented for unanimous sharing of compiled data as approved by the IRB of the college at SPHMMC.
Ethical Consideration:
The final proposal of the research was presented to IRB of the college for approval of the research and written approval was obtained. After the approval, the research was conducted in AaBET hospital without using patient’s name, staff’s name or potential identifiers. Instead, medical record numbers were used on data collection forms to ensure confidentiality.
Acknowledgement
We would like to thank the data collectors and study participants
Authors’ contributions
MT, EN: conceptualized the research problem, designed the study, conducted fieldwork, collected and data analyzed, and drafted the manuscript. MK, YW: was involved in conceptualization, preparing the research proposal, and revising the final manuscript. All authors of the manuscript have read and agreed to its content.
Funding
No funding
Competing interest
All authors read and approved the final manuscript. The authors declare that they have no competing interests.
Availability of Data and Materials
The datasets used in the current study or data collection tool are available from the corresponding author with a reasonable request.
References
-
Stewart S, Robertson C, Pan J, Kennedy S, Dancer S, Haahr L, et al. Epidemiology of healthcare-associated infection reported from a hospital-wide incidence study: considerations for infection prevention and control planning. The Journal of hospital infection. 2021;114:10-22.
--> -
Meybodi MME, Foroushani AR, Zolfaghari M, Abdollahi A, Alipour A, Mohammadnejad E, et al. Antimicrobial resistance pattern in healthcare-associated infections: investigation of in-hospital risk factors. Iranian journal of microbiology. 2021;13(2):178-82.
Publisher | Google Scholor -
.Uecker H, Bonhoeffer S. Antibiotic treatment protocols revisited: the challenges of a conclusive assessment by mathematical modelling. Journal of the Royal Society, Interface. 2021;18(181):20210308.
Publisher | Google Scholor -
Yehuala3 WWYAKFM. Point prevalence of hospital-acquired infections in two teaching hospitals of Amhara region in Ethiopia. 23 August 2016.
Publisher | Google Scholor -
Richard Smith professor of health system economics 1 J. The true cost of antimicrobial resistance.BMJ 2013;346:f1493 doi: 10.1136/bmj.f1493 (Published 11 March 2013).
Publisher | Google Scholor -
Gebretekle GB, Mariam DH, Mac S, Abebe W, Alemayehu T, Degu WA, et al. Cost–utility analysis of antimicrobial stewardship programme at a tertiary teaching hospital in Ethiopia. BMJ open. 2021;11(12):e047515.
Publisher | Google Scholor -
Labi AK, Obeng-Nkrumah N, Dayie N, Egyir B, Sampane-Donkor E, Newman MJ, et al. Antimicrobial use in hospitalized patients: a multicentre point prevalence survey across seven hospitals in Ghana. JAC-antimicrobial resistance. 2021;3(3):dlab087
Publisher | Google Scholor -
Walelegn Worku Yallew1* AK, Feleke Moges Yehuala3. Risk factors for hospital-acquired infections in teaching hospitals of Amhara regional state, Ethiopia: A matched-case control study. July 18, 2017.
--> -
Hagos A, Gedif T. Epidemiology of antibacterial drug resistance in northern Ethiopia. Ethiopian Pharmaceutical Journal. 2009(Vol. 27 No. 1 (2009)).
Publisher | Google Scholor -
.Qi Chen1 DL, Claudia Beiersmann1, Florian Neuhann1,3,, Babak Moazen1, Guangyu Lu5,* and Olaf Müller1,*. Risk factors for antibiotic resistance development in healthcare settings in China: a systematic review. Cambridge University Press. 24 May 2021.
Publisher | Google Scholor -
john E. McGowan J. Antimicrobial Resistance in Hospital Organisms and Its Relation to Antibiotic Use. REVIEWS OF INFECTIOUS DISEASES VOL 5, NO6 NOVEMBER-DECEMBER 1983. 1983.
Publisher | Google Scholor -
Lakbar I, Medam S, Ronfle R, Cassir N, Delamarre L, Hammad E, et al. Association between mortality and highly antimicrobial-resistant bacteria in intensive care unit-acquired pneumonia. Scientific reports. 2021;11(1):16497.
Publisher | Google Scholor -
Onuzo CN, Sefogah PE, Nuamah MA, Ntumy M, Osei MM, Nkyekyer K. Surgical site infections following caesarean sections in the largest teaching hospital in Ghana. Infection prevention in practice. 2022;4(2):100203.
Publisher | Google Scholor -
WHO. Antimicrobial Resistance: WHO; 2021 [cited 2021 17 November 2021]. Available from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
--> -
Avershina E, Shapovalova V, Shipulin G. Fighting Antibiotic Resistance in Hospital-Acquired Infections: Current State and Emerging Technologies in Disease Prevention, Diagnostics and Therapy. Frontiers in microbiology. 2021;12:707330.
Publisher | Google Scholor -
Aleksa Despotovic MDa, Branko Milosevic MD, PhD b,c, Ivana Milosevic MD, PhD c,d, Nikola Mitrovic MD, PhD b,c, Andja Cirkovic MDe, Snezana Jovanovic MDf, Goran Stevanovic MD, PhD c,g. Hospital-acquired infections in the adult intensive care unit—Epidemiology, antimicrobial resistance patterns, and risk factors for acquisition and mortality. American Journal of Infection Control. 2020.
Publisher | Google Scholor -
Massongo M, Ngando L, Pefura Yone EW, A NZ, Mbanzouen W, Fonkoua MC, et al. Trends of Antibacterial Resistance at the National Reference Laboratory in Cameroon: Comparison of the Situation between 2010 and 2017. BioMed research international. 2021;2021:9957112.
Publisher | Google Scholor -
Frederick K. WangaiID1 MMM, Godfrey N. Lule1‡,EmmaM. Karari1‡,, Marybeth C. MaritimID1‡ WGJ, Beatrice Museve3, Antony Kuria3. Bridging antimicrobial resistance knowledge
Publisher | Google Scholor -
.Kiros T, Damtie S, Eyayu T, Tiruneh T, Hailemichael W, Workineh L. Bacterial Pathogens and Their Antimicrobial Resistance Patterns of Inanimate Surfaces and Equipment in Ethiopia: A Systematic Review and Meta-analysis. BioMed research international. 2021;2021:5519847.
Publisher | Google Scholor -
.Beyene G, Tsegaye W. Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in jimma university specialized hospital, southwest ethiopia. Ethiopian journal of health sciences. 2011;21(2):141-6.
Publisher | Google Scholor -
Ibrahim RA, Teshal AM, Dinku SF, Abera NA, Negeri AA, Desta FG, et al. Antimicrobial resistance surveillance in Ethiopia: Implementation experiences and lessons learned. Afr J Lab Med. 2018;7(2):770-.
Publisher | Google Scholor -
Yallew WW, Kumie A, Yehuala FM. Point prevalence of hospital-acquired infections in two teaching hospitals of Amhara region in Ethiopia. Drug, healthcare and patient safety. 2016;8:71-6.
Publisher | Google Scholor -
Berhe DF, Beyene GT, Seyoum B, Gebre M, Haile K, Tsegaye M, et al. Prevalence of antimicrobial resistance and its clinical implications in Ethiopia: a systematic review. Antimicrobial Resistance & Infection Control. 2021;10(1):168.
Publisher | Google Scholor -
Mengesha Y MB, Moges G. . Assessment of Public Awareness, Attitude, and Practice Regarding Antibiotic Resistance in Kemissie Town, Northeast Ethiopia: Community-Based Cross-Sectional Study.; Infect Drug Resist. 2020;13:3783-3789.
Publisher | Google Scholor -
Darkwa EO, Djagbletey R, Sottie D, Owoo C, Vanderpuye N, Essuman R, (2018). Serum nitric oxide levels in healthy pregnant women: a case-control study in a tertiary facility in Ghana. Matern Health Neonotol Perinatol 4(3):1-
Publisher | Google Scholor -
Tabassum H, Al-Jameil N, Ali MN, Khan FA, Al-Rashed M(2015).. Status of serum electrolytes in preeclamptic pregnant women of Riyadh, Saudi Arabia. Biomed Res;26:219-24
Publisher | Google Scholor -
Akhtar S, Begum S, Ferdousi S(2011).. Calcium and zinc deficiency in pre-eclamptic women. J Bangldesh Soc Physiol 6:94-9.
Publisher | Google Scholor
